

FULL STORY
Getting Personal: The Coming Era of Individualized Medicine
Physicians have long known that no two patients are the same. Hippocrates, for example, believed that illness stemmed from imbalances of the four humors (blood, phlegm and black and yellow bile), which varied from person to person.

This article originally appeared in the Summer/Fall issue of Einstein magazine.Only in the twentieth century—following the work of Gregor Mendel, James Watson and Francis Crick and others—was it established that our individuality and susceptibility to certain diseases are rooted in our genomes—our full complement of genetic material. More recently, we’ve learned that our epigenomes—consisting of the thousands of molecules that bind to genes and control their expression—also affect our health. Since our epigenomes are influenced by environmental factors such as diet, stress, cigarette smoking and physical activity, it has now become clear that nature and nurture—our genetic and our epigenetic profiles—are interacting to make us biologically unique.
This knowledge is creating a new medical era in which healthcare is based not on what works for most patients but on what is ideal for a subgroup of patients or even particular patients, based on their genetic or epigenetic profiles. Variously referred to as individualized, personalized or precision medicine, the field was made possible in large part by the 13-year human genome–mapping project, completed at a cost of $3 billion in 2001. Since then, the cost of sequencing all or part of a person’s genome has plunged dramatically, along with the time required for the task.
"Personalized medicine is… the tailoring of medical treatments to the individual characteristics of each patient, and the ability to classify individuals into subpopulations based on their susceptibility to a particular disease or their responses to a specific treatment. Personalized medicine therefore has the potential to optimize targeted delivery and dosing of treatments so patients can receive the most benefit with the least amount of risk, cutting out the difficulties of the current trial-and-error process many patients endure to find the correct drug and dose to treat a condition.”"
– President’s Council of Advisors on Science and Technology (2008)
In the near future, “whole-genome sequencing” is expected to cost as little as $1,000—comparable to a routine MRI or colonoscopy—and take just a single day.
“Genomic sequencing is a technology whose time has come,” says Harry Ostrer, M.D., professor of pathology, of genetics and of pediatrics at Einstein and director of genetic and genomic testing in clinical pathology at Montefiore, the University Hospital and academic medical center for Einstein. “It will be transformative. We will be able to predict a patient’s risk for disease,the course of the disease, whether the patient will respond to this or that therapy and whether the patient will experience toxic side effects.”
Dr. Ostrer notes that personalized medicine is already bolstering cancer treatment for patients whose tumors are driven by specific gene mutations that can be precisely targeted by drugs. But don’t expect it to totally transform medical therapy any time soon. “It’s going to be a sequential rollout—one disease, one test, one therapy at a time,” he cautions. “We would be giving you risk assessments for very specific conditions.”
Although personalized medicine will one day be based on whole-genome sequencing, Dr. Ostrer notes the large gap that now exists between the massive amount of data generated from wholegenome sequencing and our ability to interpret those data.

Harry Ostrer, M.D., Professor of pathology, of genetics and of pediatrics“To say, at this point, that we are going to be able to sequence your whole genome and then give you some sort of meaningful analysis about your overall disease risk is a great exaggeration,” cautions Dr. Ostrer. “The science for that is not there yet.”
Others at Einstein agree. “We have this explosion of information about the genome and epigenome, but we don’t know what is important and what is not,” adds Michael B. Prystowsky, M.D., Ph.D., professor and chair of pathology at Einstein and at Montefiore Medical Center, who studies biomarkers that can distinguish between aggressive and nonaggressive head and neck cancers. “Bioinformatics—the processing of all this information and figuring out what it means and how it applies to the individual—is going to take a lot of work.”
Einstein’s department of genetics is tackling the major challenge of managing and analyzing the huge amount of data from their gene-sequencing work. “We anticipated this problem back in 2009 when we started using these new sequencing technologies,” says John M. Greally, M.B., B.Ch., Ph.D., who heads the division of computational genetics in the department of genetics. “People were puzzled as to why we were investing so heavily in high-performance computing and software development at the time, but we have seen it pay off since,” says Dr. Greally, also professor of genetics, of medicine (hematology) and of pediatrics, the Ruth L. and David S. Gottesman Faculty Scholar for Epigenomics, and attending physician in pediatrics at The Children’s Hospital at Montefiore.
Jan Vijg, Ph.D., professor and chair of genetics, agrees. “Our sequencing analysis capabilities are seen as a model for other institutions,” says Dr. Vijg, also professor of ophthalmology and visual sciences and the Lola and Saul Kramer Chair in Molecular Genetics. “Especially here in the Bronx, with its extraordinarily diverse population, we need to be particularly good at analyzing genetic data and tailoring our findings to people so that they can receive the optimal benefits of personalized medicine.”
Researchers across the Einstein campus are working in the realm of personalized medicine and genetic profiling, discovering—to paraphrase Leo Tolstoy—how “all unhealthy patients are unhealthy in their own way,” and translating those findings into individualized diagnostics and therapeutics. We describe the work of eight of those scientists in the following pages.
Individualized Medicine: Cancer
Tailoring Breast Cancer Therapy to Gene Expression
 In theory, there’s a simple algorithm for treating women with early-stage breast cancer: If the cancer is unlikely to recur locally or spread to other parts of the body, then surgical excision of the tumor, followed by radiation and hormonal therapy, is sufficient. But if recurrence or spread is likely, then additional treatment in the form of chemotherapy is warranted.
In theory, there’s a simple algorithm for treating women with early-stage breast cancer: If the cancer is unlikely to recur locally or spread to other parts of the body, then surgical excision of the tumor, followed by radiation and hormonal therapy, is sufficient. But if recurrence or spread is likely, then additional treatment in the form of chemotherapy is warranted.
Unfortunately, standard diagnostic criteria such as tumor size and tumor grade don’t reliably distinguish between the two types of breast cancer—the reason current treatment guidelines recommend erring on the side of caution and treating most breast cancer patients aggressively.
“While this approach ensures that most women with high-risk cancers get Joseph A. Sparano, M.D., Professor of medicine (oncology) and of obstetrics & gynecology and women’s health treated, it also means that most women with low-risk disease are overtreated and unnecessarily exposed to potentially toxic therapies,” says breast cancer specialist Joseph A. Sparano, M.D., professor of medicine (oncology) and of obstetrics & gynecology and women’s health at Einstein, director of the Clinical Trials Office at the Albert Einstein Cancer Center and chief of the section of breast medical oncology at Montefiore Medical Center. He and other Einstein researchers are searching for new tools to predict a breast cancer’s course so that doctors can deliver the right therapy to each patient.
One promising tool is called Oncotype DX. Developed by Genomic Health, a California-based biotechnology firm, Oncotype DX analyzes the expression of 21 breast tumor genes to come up with a “recurrence score” ranging from 1 to 100, with a low score associated with a low risk for recurrent cancer. The test is specifically geared toward women with estrogenreceptor-positive tumors (i.e., tumors that respond to hormonal therapy), which account for about two-thirds of all breast cancers.

Joseph A. Sparano, M.D., Professor of medicine (oncology) and of obstetrics & gynecology and women’s healthClinical trials of Oncotype DX had found that about 75 percent of patients had either a low or high score, while 25 percent had an intermediate score. Patients with low scores can be effectively treated with hormonal therapy alone, while those with high scores need chemotherapy in addition to hormone therapy to have a high likelihood of a cure. When the test started being used in clinical practice, most patients were expected to fall into either the high or low range—but the opposite turned out to be true.
“After the test was put into widespread practice, in 2005, about 65 percent of patients had scores in the intermediate range,” says Dr. Sparano. “That was because clinicians were using the test mainly on patients who met clinical guidelines pointing to chemotherapy but also had favorable or intermediate tumor characteristics. We didn’t know what therapy to recommend for women in this gray zone and needed a large-scale clinical trial to find out.”
Dr. Sparano and his fellow researchers got their wish. In 2006, the National Cancer Institute sponsored a clinical trial called TAILORx, or Trial Assigning Individualized Options for Treatment (Rx), to address several issues related to Oncotype DX—most notably, the best therapy for women with intermediate recurrence scores. A noted authority on clinical trials, Dr. Sparano was tapped to lead the study on behalf of the coordinating body, the Eastern Cooperative Oncology Group (ECOG), where he was co-chair of the breast committee.
TAILORx recruited about 10,000 patients at 900 sites in the United States, Canada, Europe, South America and Australia. All women with a midrange recurrence score of 11 to 25—about 6,900 in total—had surgery and then were randomly assigned to receive hormonal therapy or hormonal therapy plus chemotherapy. The remaining 3,100 women underwent surgery and then were assigned to hormonal therapy if the recurrence score was low and to chemotherapy plus hormonal therapy if the recurrence score was high.
Women enrolled in the trial will be followed for up to 25 years. Tissue collected in the study will be stored for use in future studies to learn more about breast cancer and to evaluate, and potentially refine, diagnostic tests that might prove even more useful than the Oncotype test for guiding treatment decisions.
“Developing these new tests marks an important first step towards meeting our goal of providing the right treatment for the right patient at the right time.”
TAILORx met its goal for accruing patients in October 2010, and Dr. Sparano and his colleagues are awaiting the results, which are not expected to be available until 2016. Those results could dramatically affect the care of tens of thousands of breast cancer patients by sparing them chemotherapy that would otherwise have been recommended for them.
Dr. Sparano has also worked with ECOG and Genomics Health to develop and validate a test for predicting the fate of ductal carcinoma in situ (DCIS), a condition in which abnormal cells are found within a breast duct.
Most cases of DCIS are cured with surgery plus radiation, but a significant number can also be cured using surgery alone. In some cases, the DCIS can transform into a more aggressive form of cancer that can spread to other organs, making it very tricky to figure out the best treatment option for an individual patient. The new test, called the “DCIS score,” measures the expression of 12 of the same 21 genes in the Oncotype DX test but uses a different formula to calculate the score. The study carried out by Dr. Sparano’s team found that the DCIS score was able to identify which patients treated with surgery alone were more likely to have a recurrence—especially recurrence with a lethal form of breast cancer.
“This test can help us make more- informed decisions about the optimal treatment for our patients,” says Dr. Sparano. “Developing these new tests marks an important first step towards meeting our ultimate goal of providing the right treatment for the right patient at the right time.”
Predicting Breast Cancer Metastasis
Another group of Einstein researchers has devised a test that predicts whether a breast tumor will metastasize, so women with nonaggressive forms of the disease can be spared from undergoing unnecessary chemotherapy.
Several years ago, a team led by John S. Condeelis, Ph.D., professor and cochair of anatomy and structural biology, co-director of the Gruss Lipper Biophotonics Center and the Judith and Burton P. Resnick Chair in Translational Research, found that breast cancers metastasize, or spread, only when a specific trio of cells co-localize at a microanatomic site on a blood vessel. This complex of cells consists of an endothelial cell (a type of cell that lines the blood vessels), a perivascular macrophage (a type of immune cell found near blood vessels) and a tumor cell that produces a protein called Mena, which enhances a cancer cell’s invasiveness. A site with these three cells constitutes what is called a tumor microenvironment of metastasis, or TMEM.

John S. Condeelis, Ph.D., Professor and co-chair of anatomy and structural biologyThen a team involving Dr. Condeelis’ group and investigators from New York–Presbyterian/Weill Cornell and MIT developed a tissue test to detect the presence and density of TMEMs. The test consisted of a triple immunostain containing antibodies to the three cell types. A high number of TMEMs in a tissue sample means that the tumor is likely to metastasize or has already done so.
To assess the test’s accuracy, it was tried on surgical tissue samples from 30 patients with metastatic breast cancer and 30 patients with localized breast cancer, all of whom had been followed for at least five years. The resulting immunostains were evaluated by two pathologists who were not aware of the patients’ clinical outcomes.
The analysis confirmed that TMEM density is significantly higher in patients who had developed metastatic breast cancer than in those who had localized disease. For every 10-unit increase in TMEM density, the risk for metastatic disease doubled.
While the new test promises to reduce overtreatment of breast cancer, it could reduce undertreatment as well. “There are some patients with grade 1 breast cancer who ultimately develop metastatic disease,” says Dr. Condeelis. “By measuring TMEM counts, we could identify those people and treat them appropriately.”
The current TMEM test requires surgical tissue, but the researchers are working on a simpler, blood-based version. “It could be part of a regular checkup, especially for women with a strong family history of the disease,” says Dr. Condeelis, also scientific director of Einstein’s Analytical Imaging Facility.
Einstein researchers are now conducting a study, involving some 500 women, to validate the TMEM test. This effort is being led by Thomas E. Rohan, M.D., Ph.D., professor and chair of epidemiology & population health at Einstein and Montefiore, and the Atran Foundation Chair in Social Medicine at Einstein.
A Crystal Ball for Cervical Cancer
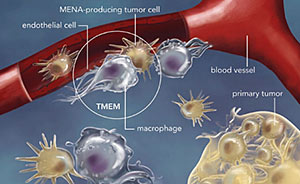
Metastasis requires the presence of three cells in the same microanatomic site: a tumor cell that produces the protein Mena; a macrophage (a cell that guides tumor cells to blood vessels); and a blood-vessel endothelial cell. The presence of three such cells in contact with each other is called a tumor microenvironment of metastasis, or TMEM, which is depicted within the circle in this illustration.A century ago, cervical cancer was the number one cancer killer among American women. Today, it’s not even among the top ten, thanks to widespread use of the Papanicolaou test (better known as the Pap smear), which can detect potentially precancerous changes in the cervical lining. Now that there’s a vaccine for human papilloma virus (HPV)—the underlying cause of virtually all cervical cancers—fewer women are likely to develop the disease in the years ahead.
Despite this progress, the clinical management of women at risk for cervical cancer is not ideal. Studies have shown that only one in ten women with persistent HPV infection and early precancerous cervical lesions will develop more-serious disease. “But there’s no way to determine exactly which one of those ten will actually progress,” explains Mark H. Einstein, M.D., M.S., associate professor of obstetrics & gynecology and women’s health and of epidemiology & population health at Einstein and director of clinical research for women’s health and gynecologic oncology at Montefiore.
As a precaution, clinicians typically advise all women with persistent HPV infection and suspicious cervical lesions to get regular follow-up tests, a scenario that can go on for years. “This is not trivial,” says Dr. Einstein. “Each year, about 3 percent of women have equivocal Pap smears, which means millions of women who must endure extra testing, not to mention the added anxiety. We need a crystal ball to predict which early lesions will regress and which will progress.”

Mark H. Einstein, M.D., M.S., Associate professor of obstetrics & gynecology and women’s health and of epidemiology & population health Or perhaps the scientific equivalent of a crystal ball: a test for detecting molecular patterns hidden in a precancerous cervical lesion that telegraph the eventual onset of cancer. Dr. Einstein and his colleagues suspect that cervical cancer starts with an epigenetic process called DNA methylation, which involves molecules known as methyl (CH3) groups that bind to and silence certain genes that regulate normal cellular processes. DNA methylation occurs all the time in cells, but when it affects tumor-suppressor genes (which, as their name implies, help keep cancerous cells in check), cancer can develop. If gene silencing helps spur cervical cancer, it should be possible to distinguish benign lesions from potentially dangerous ones by looking for differences in the epigenetic patterns inside their cells. To test his hypothesis, Dr. Einstein has been studying 100 patients with persistent HPV infection and early precancerous cervical lesions. The patients—recruited from Montefiore Medical Center in the Bronx, home to a population at especially high risk for cervical cancer—are being examined every six months for two years, and their lesions are analyzed for epigenetic changes affecting tumorsuppressor genes.
“We hope that knowledge from this study will eventually allow us to triage women who need aggressive treatment versus those who can be followed safely and conservatively without treatment,” says Dr. Einstein.
A Leg Up on Head and Neck Cancer
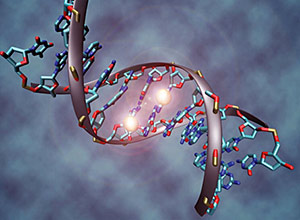
Illustration of a DNA molecule that is methylated at the two center cytosine bases. DNA methylation plays an important role in normal development and in disease.Head and neck cancers are the sixth most common malignancy among men worldwide and one of the deadliest: only half of such patients are still alive more than five years after diagnosis. This survival rate hasn’t budged in 40 years, partly because physicians have no way of knowing which tumors are likely to spread and therefore warrant aggressive treatments such as the surgical removal of surrounding lymph nodes.
Earlier this year, researchers at Einstein and Montefiore, its clinical partner, found a biomarker in head and neck cancers that appears to predict whether a patient’s tumor will be life-threatening. The biomarker is especially promising because it can indicate the risk level immediately after the cancer is diagnosed, notes the study’s co-leader, Geoffrey J. Childs, Ph.D., professor of pathology.
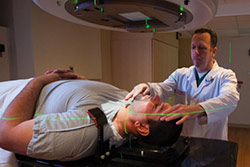
With the aid of a technician, a head-and-neck cancer patient undergoes radiation therapy at Montefiore Medical Center.Dr. Childs’ study involved 123 head and neck cancer patients. He and his colleagues took tissue samples from the tumors and nearby healthy tissue and measured levels of 736 members of a class of RNA molecules known as microRNAs (miRNAs). Of the miRNAs measured, one in particular—miR-375—stood out for being the most downregulated (i.e., expressed at low levels) in cancerous tissue compared with its levels in adjacent normal tissue.
The researchers ranked the patients according to how extreme the difference was between the miR-375 in their tumors and in surrounding healthy tissue. All of the patients were then followed throughout the course of their illness. MiR-375 proved to be highly useful for predicting disease outcome. The patients for whom the difference between their tumors and normaltissue miR-375 levels was most extreme (i.e., the one-fourth of patients with the lowest tumor-to-healthy-tissue miRNA ratios) were nearly 13 times more likely to die or 9 times more likely to experience distant spread (metastasis) of their cancer, compared to patients with higher miR-375 ratios.
“We hope that miR-375 will become part of a laboratory test to determine which patients have potentially lethal tumors and therefore should be treated aggressively following initial diagnosis,” says Dr. Childs, whose study was published in the American Journal of Pathology. “And for patients with less-threatening disease, such a test could spare them the potentially poor quality of life that can result from aggressive surgery.”
The Right Drug for the Right Colon Cancer Patient
Over the last decade or so, treatment advances have added a full year to the median survival of patients with metastatic colon cancer. Much of the credit goes to a new class of drugs called EGFR (epidermal growth factor receptor) inhibitors. They block EGFR, a cell-surface receptor that fuels cell growth and proliferation and is abundantly present on the surface of colon cancer cells (and is found in other types of cancer as well).

Sanjay Goel, M.D., M.S., Associate professor of medicine (oncology)Unfortunately, a large percentage of colon cancer patients do not respond to EGFR inhibitors. The cancer’s response to these drugs depends on a gene called K-ras: tumors with a mutated form of K-ras are drug resistant. But something else must also be counteracting these drugs.
“Even if you exclude colon cancer patients with mutant K-ras genes, you still find that only about half of colon cancers respond to EGFR inhibitors,” says Sanjay Goel, M.D., M.S., associate professor of medicine (oncology) at Einstein and attending physician in oncology at Montefiore. In laboratory studies of colon cancer cells from 22 patients, Dr. Goel and his colleagues discovered two other genetic anomalies—mutations to the genes PIK3CA and PTEN—that reduce a tumor’s sensitivity to EGFR inhibitors.
Moving from the lab to a study of 76 colon cancer patients, the researchers found that the presence of the two mutations reliably predicts a poor response to EGFR inhibitors, as they reported this year in Clinical Colorectal Cancer. Einstein and Montefiore have since filed and received a patent on the use of PIK3CA mutations to predict whether or not patients will benefit from EGFR inhibitors. The patent has been licensed for development by Transgenomic, Inc., an Omaha, NE, biotechnology company.
The PIK3CA test could spare a fair number of patients from potentially dangerous treatments that won’t help them. “EGFR inhibitors can be quite toxic, with side effects ranging from skin rashes to diarrhea,” says Dr. Goel. The test would also reduce healthcare expenditures, he notes, since taking these drugs can cost thousands of dollars a month.
Individualized Medicine: Heart Disease
Minimizing Heart Attack Damage
 A cut to the skin is usually benign: a clot forms, and maybe a small scar, but most evidence of the wound soon vanishes. Unfortunately, this minor, everyday miracle of repair and regeneration doesn’t extend to the heart. After a heart attack, the body can do little to rejuvenate damaged heart muscle. Scars form but never disappear, limiting heart function. Over time, the heart begins to change its architecture in an attempt to wring more pumping power from the weakened organ. This process, known as remodeling, may help in the short term but ultimately can lead to a host of health issues—most notably heart failure.
A cut to the skin is usually benign: a clot forms, and maybe a small scar, but most evidence of the wound soon vanishes. Unfortunately, this minor, everyday miracle of repair and regeneration doesn’t extend to the heart. After a heart attack, the body can do little to rejuvenate damaged heart muscle. Scars form but never disappear, limiting heart function. Over time, the heart begins to change its architecture in an attempt to wring more pumping power from the weakened organ. This process, known as remodeling, may help in the short term but ultimately can lead to a host of health issues—most notably heart failure.
So the cardiologist’s first task when treating a heart attack patient is to minimize heart muscle damage, and job number two is to limit remodeling. Over the years, doctors have made great strides in achieving the former but not the latter, says Nikolaos G. Frangogiannis, M.D., professor of medicine (cardiology) and the Edmond J. Safra/Republic National Bank of New York Chair in Cardiovascular Medicine.
The problem, Dr. Frangogiannis explains, is that remodeling can take two different forms: dilated remodeling (in which the heart muscle stretches and thins) or hypertrophic remodeling (in which the walls of the heart’s ventricles thicken and stiffen). Each form requires a different treatment approach, but it’s impossible to know which disease path the heart will follow before harmful changes occur. And once remodeling starts, it’s hard to stop or reverse.

Nikolaos Frangogiannis, M.D., Professor of medicine (cardiology)“We end up treating all heart attack patients pretty much the same way,” says Dr. Frangogiannis, who studies the molecular signals that orchestrate cardiac healing after a heart attack. “Ideally, we would have a panel of biomarkers that, together with a patient’s genetic background, clinical history and other factors, could tell us early on whether to treat for dilated or hypertrophic remodeling.”
Dr. Frangogiannis may have identified one such biomarker. In a series of studies published in Circulation, Circulation Research and the Journal of Molecular and Cellular Cardiology, he found that transforming growth factor beta (TGF-beta), a molecule involved in regulating immune and inflammatory responses as well as in tissue repair, is elevated after a heart attack and increases heart muscle stiffness. His findings suggest that TGF-beta causes connective tissue cells called fibroblasts to pump out excess collagen, which in turn leads to fibrosis and to thickening and stiffening of the heart muscle.
Dr. Frangogiannis hopes that a test will be developed (an imaging test or one that relies on genetic profiling, for example) to identify patients who have high levels of TGF-beta after a heart attack and are therefore at risk for hypertrophic remodeling. Such a test could lead to personalized post–heart attack treatment strategies that would curb the expression of TGF-beta as the heart heals.
Cardiogenetics in Action

Thomas V. McDonald, M.D., Professor of medicine (cardiology) and of molecular pharmacologyMany individualized diagnostics and therapeutics have already found their way into everyday clinical practice. The recent case of Adam (not his real name), a young boy who was treated at The Children’s Hospital at Montefiore, is a perfect example.
After fainting several times, Adam was diagnosed with long QT syndrome, a type of arrhythmia that can cause fast, chaotic heartbeats, with potentially fatal consequences. In the past, the boy would probably have been given the standard anti-arrhythmia therapy, a beta blocker. His doctors might also have recommended an implantable cardiodefibrillator (ICD), a small device that can instantly detect and correct abnormal heart rhythms.
Since implanting an ICD requires surgery and carries some risks, it is usually reserved for patients with dangerous arrhythmias or a lethal family history of the disease. But Adam was adopted and his family’s history was unknown, so the risk-benefit ratio of implanting an ICD was difficult to assess. Fortunately, Adam’s Montefiore doctors were able to find exactly what therapies he needed, no more and no less.
“We now know many of the genetic mutations that can cause long QT,” explains one of his caregivers, Thomas V. McDonald, M.D., professor of medicine (cardiology) and of molecular pharmacology at Einstein, attending cardiologist at Montefiore and co-director of the Einstein-Montefiore Cardiogenetics Clinic. “So, as a matter of routine, we sent a sample of Adam’s DNA to the lab for genetic testing.”
The test revealed a genetic variant never before seen—a result that would put most cardiologists back at square one. But Dr. McDonald’s lab was equipped to delve deeper into Adam’s DNA anomalies using sophisticated genetic sleuthing. Then his team recreated that mutation in a cellular model and determined its physiological effect.
“We found that Adam’s mutation wasn’t a severely deleterious variant, so an ICD wasn’t necessary,” he says. “We were able to reassure his family that he has little risk of sudden cardiac death."
What’s more, Dr. McDonald was able to recommend appropriate treatment for Adam’s particular arrhythmia risk. All genetic mutations that cause long QT do so by disturbing ion channels in the heart. However, the various mutations affect different channels in different ways and thus require different remedies.
“In Adam’s case, the genetic variation caused a gain of function in his heart’s potassium channels, which is best treated with beta blockers,” says Dr. McDonald, a basic researcher as well as a clinician. “If, on the other hand, his potassium channels were underactive, we would be looking at different drugs.”
Individualized Medicine: Diabetes
Predicting Type 2 Diabetes Risk
If you’re overweight or lead a sedentary lifestyle, you face an elevated risk of developing type 2 (adult onset) diabetes. But a number of other factors also influence whether someone will become diabetic.
“We know, for example, that obesity is a major risk factor for diabetes. But some overweight individuals don’t develop diabetes, while some thin people do,” explains Howard D. Strickler, M.D., M.P.H., professor of epidemiology & population health. In his quest to understand the molecular underpinnings of diabetes, Dr. Strickler may have identified a more accurate way to estimate someone’s risk for developing diabetes, long before the first signs and symptoms emerge.
“Obesity is a major risk factor for diabetes. But some overweight individuals don’t develop diabetes, while some thin people do.”

Howard Strickler, M.D., M.P.H., Professor of epidemiology & population health, pictured here with colleague Mimi Kim, Sc.D., professor and head of the division of biostatistics in the department of epidemiology & population healthDr. Strickler has focused on the IGF axis—a group of proteins that includes insulin-like growth factor-1 (IGF-1). IGF-1 earned its name because it shares some biological effects with insulin (the hormone that regulates blood glucose levels) but has a greater effect on growth than insulin does. The IGF axis also includes six IGF binding proteins, or IGFBPs, that may have strong effects independent of IGF-1.
Researchers have hypothesized that the IGF axis may influence risk for developing diabetes—an idea supported by laboratory and mouse studies, and a few initial human studies. Now, in the largest and most comprehensive prospective study of its kind, Dr. Strickler and his colleagues have found that levels of certain IGF-axis components found in blood are associated with a greatly increased risk for developing diabetes a decade or more later.
The researchers analyzed levels of IGF-1, IGFBP-1, IGFBP-2 and IGFBP-3 in blood taken from 742 women in the Nurses’ Health Study who years later developed type 2 diabetes, as well as a similar number of women in the study who did not develop diabetes. None of the women had any signs or symptoms of diabetes at the time their blood samples were taken. The median time between the taking of blood samples and diabetes onset was nine years.

A patient with diabetes obtains blood for measuring his blood glucose level. Research at Einstein may help people learn they’re at risk for diabetes long before it occurs.Each component of the IGF axis analyzed (IGF-1 and IGFBP-1, -2 and -3) had a significant independent association with diabetes risk—most notably IGFBP-1 and -2. Women with the lowest levels of IGFBP-1 had a threefold increase in risk for diabetes, compared with those with the highest levels of the protein. Women with the lowest levels of IGFBP-2 had a fivefold increase in diabetes risk compared with women with the highest levels, according to the study, published earlier this year in Diabetes.
If the findings are confirmed, adds Dr. Strickler, they could help doctors more accurately determine who is actually at risk for the disease, long before it occurs—which could give millions of diabetes-prone people ample warning and added incentive to lose weight, get more exercise and change their diets. In addition, Dr. Strickler’s studies could eventually lead to novel therapies that prevent or treat diabetes and that are specifically tailored to the individual’s unique IGF-axis signature.
Dr. Strickler cautions that it’s too early to apply these findings to clinical practice. “IGF-axis proteins have other effects, some beneficial and some not,” he notes. “We need to learn more about the connection between the IGF axis and diabetes before we recommend that people get tested for these substances, and before deciding how we can exploit the IGF-1 axis to help address diabetes.”
Posted on: Tuesday, January 15, 2013

Tablet Blog